

Host-plant relationships of 29 Mediterranean Lepidoptera species in forested ecosystems unveiled by DNA Barcoding (Insecta: Lepidoptera)
Relación de las plantas nutricias de 29 especies de Lepidoptera mediterráneas en ecosistemas forestales revelados por el Código de barras de ADN (Insecta: Lepidoptera)
Host-plant relationships of 29 Mediterranean Lepidoptera species in forested ecosystems unveiled by DNA Barcoding (Insecta: Lepidoptera)
SHILAP Revista de lepidopterología, vol. 44, núm. 175, 2016
Sociedad Hispano-Luso-Americana de Lepidopterología
Recepción: 26 Julio 2015
Aprobación: 10 Octubre 2015
Abstract: A total of 45 Lepidoptera larvae were collected in ethanol from 16 target plant species in southern Italy, requiring a few hours of field work only. Identification was performed by DNA barcoding unveiling host-plant relationships for 29 Lepidoptera species. The authors propose a larger-scale approach using this rapid and efficient method and encourage entomologists to join the team.
Keywords: Insecta, Lepidoptera, caterpillar, feeding behavior, tree species, larval identification, Italy.
Resumen: Se colectaron un total de 45 larvas de Lepidoptera en etanol de 16 especies de plantas en el sur de Italia, requiriendo sólo unas pocas horas de trabajo de campo. La identificación fue llevada a cabo con el sistema de código de barras de ADN revelando la relación con las plantas nutricias de 29 especies de Lepidoptera. Los autores proponen un enfoque a mayor escala usando este método rápido y eficaz y animando a otros entomólogos a sumarse al equipo.
Palabras clave: Insecta, Lepidoptera, oruga, comportamiento alimenticio, árbol de especies, identificación larvaria, Italia.
Introduction
Available literature on host-plant relationships of Mediterranean Lepidoptera is very scarce and feeding biology of many species is still unknown. Only a very few authentic, original data are published for host-plant relationships of Mediterranean Lepidoptera and most of them concern pest species, such as Tuta absoluta (Meyrick, 1917) (Gelechiidae) (PORTAKALDALI et al., 2013), or caterpillars found on plants of economic importance such as Euzophera bigella (Zeller, 1848) (Pyralidae) on Olea europaeaL. (SIMOGLOU et al., 2012), Lymantria dispar (Linnaeus, 1758) (Lymantriidae) on Quercus suber L. (PROTA, 1970), Cynaeda perspectalis (Walker, 1859) (Crambidae) on Buxus (KRUGER, 2008), and others. Books like those of GÓMEZ DE AIZPURUA (1985-1992, 2002-2012) on Iberian moth larvae report a lot of host-plant data, but in most cases it remains unclear if the data come from observations in nature or not. It is most likely that the vast majority comes from artificial conditions during rearing. The situation is still worse for southern Italian Lepidoptera, but an exception is the paper devoted to the lepidopteran fauna of the Quercus suber L. in Sardinia showing a large number of species feeding on this tree (PROTA, 1970). We estimate that for >90% of the putatively 4000 southern Italian Lepidoptera species (approx. 2500 Macrolepidoptera species are mentioned in PARENZAN & PORCELLI 2006; 2008 for the Italian fauna) no authentic feeding records from nature are published for the study area. We furthermore estimate that for >50% of these species no authentic feeding record from nature is reported for the whole of their distribution areas.
Few original data concern species of non-economic importance, e.g. several Microlepidoptera species which are often reared from larvae. Caryocolum siculum (Gelechiidae) was described rearing larvae on Gypsophila arrostii Guss. (Caryophyllaceae) (BELLA, 2008), Coleophora sardiniae was described rearing larvae on seeds of Genista corsica(Loisel.) DC. (Labiatae) (BALDIZZONE, 1983).
Not only in old papers devoted to European caterpillars and their food-plants (e.g. ROÜAST, 1883, 1884; HOFMANN, 1983) but also in large recent book series, such as Microlepidoptera of Europe, Lepidoptera of Europe (LERAUT, 2006-2014) and Noctuidae Europaeae it is usually impossible to trace original observations and separate them from copied and unreliable, secondary literature citations. In the book series “The Geometrid Moths of Europe” (HAUSMANN, 2001, 2004; MIRONOV, 2003; HAUSMANN & VIIDALEPP, 2012; SKOU & SIHVONEN, 2015) as well as in the“Schmetterlinge Baden-Württembergs (EBERT 1991-2005, ed.) however, there is a consistent system allowing discrimination between original (feeding) observations in nature, copied literature citations (usually copied without examining their plausibility) and data from rearing, potentially altered by the artificial conditions.
Availability of affordable data is of great importance for forestry, agriculture and ecological research mainly from a conservation point of view, but they could also be important to design environment-friendly control strategies against pests and defoliators.
The classical method to identify larvae is to rear them in captivity until the emergence of the adult and identify it through morphology. This method is time consuming and the rearing success may be quite low, especially if the hostplant is not easily available where the rearing takes place. Identification of lepidopteran larvae was successfully carried out by DNA barcoding (COI 5’) (MILLER et al., 2007; EMERY et al., 2009; GOSSNER & HAUSMANN, 2009), permitting an easy, cheap and rapid identification of species. MILLER et al. (2007) and MATHESON et al. (2008) investigated and ascertained relationships between plants and caterpillars by a method based on the DNA identification of the larval gut content, an effective but expensive and time-consuming approach, especially in larger surveys. Identification through DNA barcoding is possible even from dry skins after moulting and from empty pupal exuviae after hatching of the moths (own, original, unpublished data). In southern Italy, SCALERCIO et al. (2014) identified through DNA barcoding a larva of the endemic Hydriomena sanfilensis (Stauder, 1915) (Geometridae) feeding on Rosa canina L., the first record of a host-plant for this species. Currently, there are large-scale projects devoted to identification larvae along with their host- plants in Papua New Guinea (MILLER et al., 2013) and Costa Rica (JANZEN & HALLWACHS, 2015). Both are based on an integrative approach combining morphology, rearing and molecular techniques.
The aim of this paper was to establish host-plant relationships of caterpillars based on the identification of larvae collected on their foodplants by DNA barcoding. In this approach we assume (and in several cases observed) that the collected larvae feed on the plants on which they stay. This assumption is based on the behaviour of larvae usually resting on feeding plants during their development. Exceptions are larvae of certain species which abandon their host-plants searching for a hidden resting place during daytime and mature larvae leaving their food-plant in the last days before pupation looking for a suitable pupation site, sometimes far from their feeding plants. Only during this short timespan it is possible to collect larvae on plants different from the typical host-plant.
Material and methods
In this paper we report the results of 12 days of fieldwork in 2013 and 2014 (with a total of 21 working hours of a single person) collecting in southern Italy (Calabria and southern Basilicata regions, Table I). Larvae were beaten from plants or, sometimes, directly collected on them. Sampling from trees was performed by spreading a white sheet below the tree branches and beating their tips in order to avoid sampling of feeders on epiphytic lichens and mosses. Successively the sheets were inspected and larvae preserved in absolute alcohol. Larvae from herbaceous plants were sampled by beating or sweeping with a robust net in order to sample larvae with the net before falling down. In both cases we carefully made sure that collected specimens came from the target plant species. This was ascertained by choosing target plants isolated from other plant species. Few larvae were directly collected on feeding plants while they were feeding.
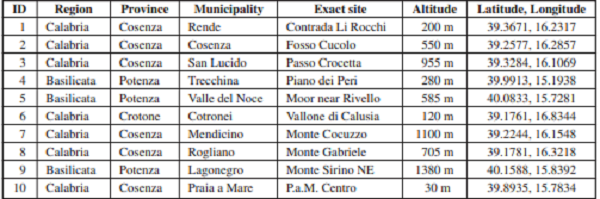
44 larvae were submitted for DNA barcoding. One additional larva was reared to the adult stage and its pupal exuvia was submitted for DNA barcoding. Vouchers are stored at the Bavarian State Collection of Zoology, Germany.
Tissue sampling was carried out by using scissors and pincers which were carefully cleaned in 100% alcohol in order to avoid contamination of samples. Tissues (one vertically cut segment) were transferred to a lysis plate under ca 0.5 ml 100% alcohol.
Tissue samples were submitted according to the standard procedures of the Canadian Centre for DNA Barcoding (CCDB) for sequencing the mitochondrial 5’ cytochrome oxidase gene, subunit 1 (COI), the standard marker for the identification of most animals. LepF1 and LepR1 were the primers used for PCR and sequencing (HAJIBABAEI et al., 2006). Sequences were blasted against the complete sequence database of BOLD data systems (RATNASINGHAM & HEBERT, 2007) in order to investigate the closest matches using the BOLD Identification Engine (http://www.boldsystems.org/index.php/ IDS_OpenIdEngine).
For nomenclature of moth names we follow the Fauna Europaea (KARSHOLT & VAN NIEUKERKEN, 2013).
Plant identification was based on morphology in most cases and carried out in the field. In some cases we collected leaves, fruits or blossoms and had the species identified by botanists specialized in the flora of southern Italy, of the Museo di Storia Naturale della Calabria e Orto Botanico, Italy. For nomenclature of plant names we follow the Flora Europaea (available online at http://rbg- web2.rbge.org.uk/FE/fe.html).
Results and Discussion
A total of 45 larvae were collected on 16 target plant species. 32 larvae belonging to 27 species were identified at species level by DNA barcoding (Tables II, III). Four additional larvae belonging to two further species could be identified to species level based on larval morphology, one to genus level. Altogether we publish host-plant relationships for 29 species, here. 13 larvae (out of 45 larvae submitted to DNA barcoding = 29%) failed to yield DNA sequences, probably because the absolute alcohol content was excessively diluted by larval fluids in too small vials, especially in the case of large caterpillars.
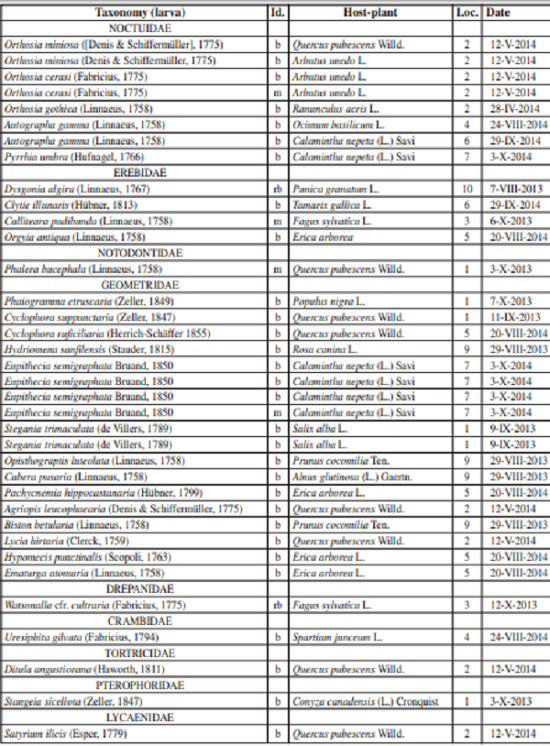
List of 36 identified larvae belonging to 29 species. Abbreviations: Id. = method of identification: b = larva identified by DNA barcoding (COI 5’, then blasted on BOLD); rb = larva reared and adult or exuvia submitted to DNA barcoding; m = morphological identification of larva; Loc. = locality code: see Table I.
For largely polyphagous species, such as Autographa gamma and Dysgonia algira, the presented records are not surprising and probably already published somewhere in literature. For little known species and for those feeding on just a few plants, it is easier to overview the published records in literature. The following records are of particular interest.
The pterophorid Stangeia siceliota was recorded on Conyza canadensis for the first time. Other known host-plants are Cistus sp., Sanguisorba sp., Inula viscosa L., Ononis natrix L. (GIELIS, 1996).
Hypomecis punctinalis (Geometridae, Ennominae) was found on Erica arboreafor the first time. Other known hostplants are Quercus, Tilia, Betula, Salix, Genista and Lonicera (REDONDO et al., 2009), furthermore Larix, Carpinus, Alnus, Berberis, Sorbus, Rubus, Prunus, Acer, Frangula, Cornus and Vaccinium (EBERT, 2003).
Phaiogramma etruscaria (Geometridae, Geometrinae) was found on Populus nigra and thus on Salicaceae for the first time. HAUSMANN (2001) characterized this species as polyphagous with a certain preference for Umbelliferae (Apiaceae), listing 14 species of herbs and shrubs, but also Quercus.
Stegania trimaculata (Geometridae, Ennominae) was found on Salix albafor the first time in nature, a plant which had been accepted only in captivity, so far (RUNGS, 1981). Usually it feeds on Populus and Alnus (FLAMIGNI et al., 2007).
A very young larva of Orthosia gothica (Noctuidae, Noctuinae) was collected on a plant of Ranunculus acris, where it was observed feeding on the petals. This plant seems to be listed as a host- plant for this moth species for the first time here, though caterpillars are known to be polyphagous on several tree species and more rarely “on various herbs” (e.g. FORSTER & WOHLFAHRT, 1980).
Furthermore, in this paper we publish the COI sequences of Clytie illunaris, Uresiphita gilvata and Stangeia siceliota for the first time.
26 out of 32 sequences blasted in BOLD received a 100% match with existing sequences on BOLD, whilst there was a small divergence of just one basepair in four other sequences and a smaller match (98.83%) for one sequence (Table III). These diverging sequences belong to Clytie illunaris, Eupithecia semigraphata and Watsonalla cfr. cultraria which resulted to be very interesting species genetically.
Clytie illunaris (Erebidae) shows at least three different haplotypes in the Mediterranean. One haplotype occurs in France and Spain, one in Israel, and two belonging to the same genetic lineage in southern Italy. Our barcoded larva just differed by one basepair from an existing Calabrian barcode on BOLD (collection specimen) and thus confirmed the presence of the diverging lineage in Calabria.
Eupithecia semigraphata (Geometridae) exhibits a high genetic diversity in its European range, especially in southern Italy where it shows at least 13 haplotypes grouping to three major lineages at larger distances. Some haplotypes seem to be restricted to Sicily and Calabria. At one of our collecting sites we found three haplotypes (BC ZSM Lep 86015/86017/86019) belonging to two different E. semigraphata-BINs (BOLD:AAB6662/ACS6376) at comparatively large distances of 1.1-1.9%. These syntopic haplotypes were found on the same Calamintha nepeta plant, suggesting that different females laid their eggs on a single plant. It might be worth testing, however, whether polymorphisms in mitochondrial genes can lead to polymorphic descendants of one single female in this species. Vulnerable plants attracting egg-laying females might be a more likely explanation than the occurrence of three mitochondrial lineages in a single egg-laying female that reached fixation in its offspring.
Perhaps the most interesting case taxonomically concerns Watsonalla cfr. cultraria (Drepanidae). The neighbor-joining tree generated from BOLD database shows specimens morphologically identified as W. cultraria placed within two different clusters. One includes specimens from the Netherlands to Macedonia, and includes the Basilicata region of southern Italy, the other (at a distance of 1.1%) includes Watsonalla specimens identified either as W. uncinula (Spain; perhaps misidentified) or W. cultraria (Tunisia and Calabria region). Apart from these two clusters there is a third cluster including W. uncinula from southern Italy and France, at distances of 1.1-1.9% from the first two and making paraphyletic the two W. cultraria clusters in some but not all tree constructions. We can hypothesize the existence of a third taxon in Spain, North Africa and southernmost peninsular Italy, but this question deserves further study.
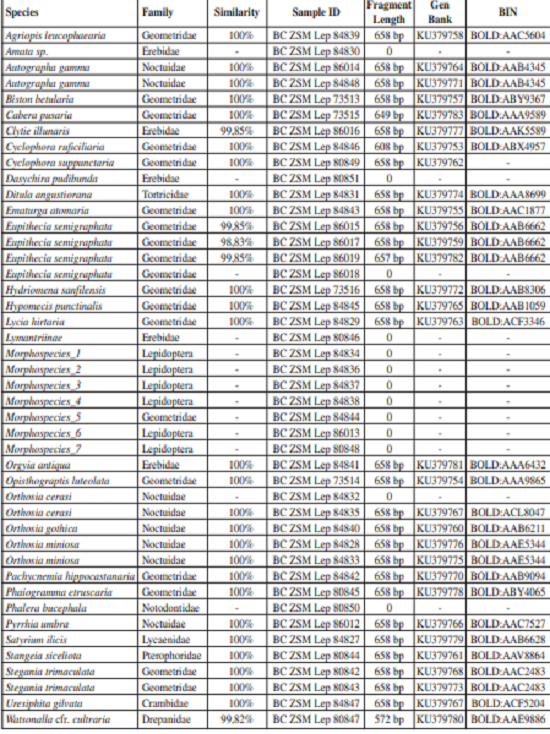
List of larvae submitted to DNA barcoding, in alphabetic order of the species name. For each sample the genetic similarity with conspecific sequences on BOLD is given, along with sample ID, fragment length, GenBank Accession number, and BIN.
Conclusions
The method used in this paper allows for the collection of many data in a few hours of field and laboratory work. The identification success was low (67.5% in this paper) compared to other studies (77.0% in GOSSNER & HAUSMANN, 2009; 97.3% in MILLER et al., 2013), but higher than the rates usually obtained by the time-consuming rearing of larvae to adults in captivity . The identification success can be increased considerably by storing the larvae in larger quantities of absolute alcohol adequate to their dimensions in order to avoid an excessive dilution.
Former critics of identification by DNA barcoding argued that species identification is hampered by missing “barcode gaps” between interspecific COI distances and intraspecific variation (WIEMERS & FIEDLER, 2007). Many recent studies (HAJIBABAEI et al., 2006; DINCA et al., 2011; HAUSMANN et al., 2011; MUTANEN et al., 2013; HUEMER et al., 2014) showed, however, that small or missing barcode gaps are very rare (and well known) in European Lepidoptera, reavealing DNA barcoding as a promising and reliable method for species identification of preimaginal stages. A major publication listing and analyzing cases of non-monophyly (barcode- sharing and barcode-splits) in European Lepidoptera is close to publication. Comprehensive DNA reference libraries of Lepidoptera are now available for a few countries of Central and Northern Europe (HAUSMANN et al., 2011; HUEMER et al., 2014). At a European level approximately 55% of the lepidopteran species have DNA barcodes at present (unpublished from own analysis of data in BOLD database). Although several species are widespread in Europe, Mediterranean countries host a peculiar fauna and DNA reference libraries for Mediterranean Lepidoptera need to be further improved.
Our approach of DNA-identification of caterpillars collected directly from their foodplants could help to fill many gaps in the knowledge of host-plant relationships of Mediterranean Lepidoptera. Furthermore we will be able to update (verify/reject) old data concerning common and widespread species which, in literature, often were transferred from other European areas without explicitly stating this. We propose to start a large-scale project on larval host-plants of Mediterranean Lepidoptera using our approach. For that purpose we encourage all interested entomologists to join our team.
Agradecimientos
We thank Carmen Gangale (Museo di Storia Naturale della Calabria e Orto Botanico, Italy) for help SS in the identification of many plant species. The genetic analyses have received considerable support from Paul D. N. Hebert and the Biodiversity Institute of Ontario (BIO) and the Canadian Centre for DNA Barcoding (CCDB University of Guelph). The data management and analysis system BOLD was provided by Sujeevan Ratnasingham. The work was financially supported by Genome Canada (Ontario Genomics Institute) in the framework of the iBOL program, WG 1.9, and by Project PON03PE_00024_1 (Ambi.Tec.Fil.Legno-AlForLab).
BIBLIOGRAPHY
BALDIZZONE, G., 1983.– Contribuzioni alla conoscenza dei Coleophoridae. XXXIII. Tre nuove specie del genere Coleophora Hübner della Sardegna. Le species del gruppo di Coleophora vulnerariae in Italia.– Entomologica, 18: 111-123.
BELLA, S., 2008.– Caryocolum siculum sp. n. (Gelechiidae), feeding on Gypsophila (Caryophyllaceae) in Sicily.– Nota lepidopterologica, 31(1): 69-75.
DINCA, V., ZAKHAROV, E. V., HEBERT, P. D. N. & VILA, R., 2011.– Complete DNA barcode reference library for a country’s butterfly fauna reveals high performance for temperate Europe.– Proceeding of the Royal Society of London, B, 278: 347-355.
EBERT, G., 1991-2005.– Die Schmetterlinge Baden-Württembergs,1-10: 5495 pp. Ulmer, Stuttgart.
EMERY, V. J., LANDRY, J. F. & ECKERT, C. G., 2009.– Combining DNA barcoding and morphological analysis to identify specialist floral parasites (Lepidoptera: Coleophoridae: Momphinae: Mompha).– Molecular Ecology Resources, ., Suppl. : 217-223.
FLAMIGNI, C., FIUMI, G. & PARENZAN, P., 2007.– Lepidotteri Eteroceri d’Italia. Geometridae Ennominae I: 382 pp. Natura Edizioni Scientifiche, Bologna.
FORSTER, W., WOHLFAHRT ,T., 1980.– Die Schmetterlinge Mitteleuropas. 4. Eulen (Noctuidae): 329 pp. Stuttgart.
GIELIS, C., 1996. Pterophoridae.– In P. HUEMER, O. KARSHOLT & L. LYNEBORG. Microlepidoptera of Europe, .: 222 pp. Apollo Books, Stenstrup.
GÓMEZ DE AIZPÚRUA, C., 1985-1992.– Biología y morfología de las orugas.– Boletín de Sanidad Vegetal, Fuera de Serie, .(1985): 240 pp., .(1986): 248 pp., .(1987): 238 pp., 4(1987): 248 pp., .(1988): 238 pp.,.(1988): 248 pp., .(1989): 224 pp., .(1990): 220 pp., .(1991): 226 pp., 10(1992): 230 pp.
GÓMEZ DE AIZPÚRUA, C., 2002-2012.– Orugas y Mariposas de Europa, .(2003): 352 pp.; .(2002): 220 pp.;.(2002): 305 pp.; .(2002): 237 pp.; .(2002): 352; .(2007): 281 pp., .(2012): 252 pp. Organismo Autónom Parques Nacionales, Madrid.
GOSSNER, M. M. & HAUSMANN, A., 2009.– DNA barcoding enables the identification of caterpillars feeding on native and alien oak (Lepidoptera: Geometridae).– Mitteilungen der Münchener Entomologische Gesellschaft, 99: 135-140.
HAJIBABAEI, M., JANZEN, D. H., BURNS, J. M., HALLWACHS, W. & HEBERT, P. D. N., 2006.– DNA barcodes distinguish species of tropical Lepidoptera.– PNAS 2006, 103: 968-971.
HAUSMANN, A. 2001.– Introduction. Archiearinae, Orthostixinae, Desmobathrinae, Alsophilinae, Geometrinae.–In A. HAUSMANN. The Geometrid Moths of Europe, .: 282 pp. Apollo Books, Stenstrup.
HAUSMANN, A., 2004.– Sterrhinae.– In A. HAUSMANN. The Geometrid Moths of Europe, .: 600 pp. Apollo Books, Stenstrup.
HAUSMANN, A., HASZPRUNAR, G., SEGERER, A. H., SPEIDEL, W., BEHOUNEK, G. & HEBERT, P. D. N., 2011.– Now DNA-barcoded: the butterflies and larger moths of Germany (Lepidoptera: Rhopalocera, Macroheterocera).– Spixiana, 34(1): 47-58.
HAUSMANN, A. & VIIDALEPP, J., 2012.– Larentiinae I.– In A. HAUSMANN. The Geometrid Moths of Europe, .: 743 pp. Apollo Books, Stenstrup.
HOFMANN, E., 1893.– Die Raupen der Gross-Schmetterlinge Europas: XXXIV + 318 pp. Verlag d. Hoffmann‘schen’ Verlagsbuchhandlung, Stuttgart.
HUEMER, P., MUTANEN, M., SEFC, K. M. & HEBERT, P. D. N., 2014.– Testing DNA Barcode Performance in 1000 Species of European Lepidoptera: Large Geographic Distances Have Small Genetic Impacts.– PLoS ONE, .(12): e115774.
JANZEN, D. H. & HALLWACHS, W., 2015.– Caterpillars, pupae, butterflies & moths of the Area de Conservación Guanacaste (ACG), northwestern Costa Rica.– Available from http://janzen.sas.upenn.edu/ (accessed 15 June 2015).
KARSHOLT, O. & VAN NIEUKERKEN E. J., 2013.– Lepidoptera. Fauna Europaea Web Service, version 2.6.2. Available from http://www.faunaeur.org.
KRUGER, E. O., 2008.– Glyphodes perspectalis (Walker, 1859) - neu fur die Fauna Europas (Lepidoptera: Crambidae).– Entomologische Zeitschrift, 118: 81-83.
LERAUT, P., 2006-2014,- Moths of Europe, .(2006): 392 pp., .(2009): 804 pp., .(2012): 599 pp., .(2014): 440 pp. NAP, Vierrières le Buisson.
MATHESON, C. D., MÜLLER, G., JUNNILA, A., VERNON, K., HAUSMANN, A., MILLER, M. A., GREENBLATT, C. & SCHLEIN, Y., 2008.– A PCR Method for Detection of Plant Meals from the Gut of Insects.– Organisms Diversity & Evolution, .: 294-303.
MILLER, M. A., MÜLLER, G. C., KRAVCHENKO, V. D., JUNNILA, A., VERNON, K. K., MATHESON, C. D. & HAUSMANN, A., 2007.– DNA-based identification of Lepidoptera larvae and plant meals from their gut content.– Russian Entomological Journal, 15(4): 427-432.
MILLER, S. E., HRCEK, J., NOVOTNY, V., WEIBLEN, G. D. & HEBERT, P. D. N., 2013.– DNA barcodes of caterpillars (Lepidoptera) from Papua New Guinea.– Proceedings of the Entomological Society of Washington, 115(1): 107-109.
MIRONOV, V., 2003.– Larentiinae II (Perizomini and Eupitheciini).– In A. HAUSMANN. The Geometrid Moths of Europe, .: 463 pp. Apollo Books, Stenstrup.
MUTANEN, M., KAILA, L. & TABELL, J., 2013.– Wide-ranging barcoding aids discovery of one-third increase of species richness in presumably well-investigated moths.– Scientific Reports, .: 290
PARENZAN, P. & PORCELLI, F., 2006.– I Macrolepidotteri Italiani. Fauna Lepidopterorum Italiae (Macrolepidoptera).– Phytophaga, 15: 5-393.
PARENZAN, P., PORCELLI, F., 2008.– I macrolepidotteri italiani. Fauna Lepidopterorum Italiae (Macrolepidoptera) - Addenda et corrigenda I.– Entomologica,15: 153-221.
PORTAKALDALI, M., ÖZTEMIZ, S. & KÜTÜK, H., 2013.– A new host plant for Tuta absoluta (Meyrick) (Lepidoptera: Gelechiidae) in Turkey.– Journal of the Entomological Research Society, 15(3): 21-24.
PROTA, R., 1970.– Contributi alla conoscenza dell’entomofauna della Quercia da sughero (Quercus suber L.). VI. Osservazioni su alcuni Lepidotteri dannosi alla Quercia da sughero (Quercus suber L.) in Sardegna.– Memoria, 30: 81 pp. Stazione Sperimentale del Sughero, Tempio Pausania.
RATNASINGHAM, S. & HEBERT, P. D. N., 2007.– BOLD: The Barcode of Life Data System (www.barcodinglife.org).– Molecular Ecology Notes, .: 355-364.
REDONDO, V. M., GASTÓN, F. J. & GIMENO, R., 2009.– Geometridae Ibericae: 361 pp. Apollo Books, Stenstrup.
ROÜAST, G., 1883.– Catalogue des chenilles Européennes connues.– Annales de la Société Linnéenne de Lyon, Nouvelle Série, 29(1882): 251-363.
ROÜAST, G., 1884.– Catalogue des chenilles Européennes connues.– Annales de la Société Linnéenne de Lyon, Nouvelle Série, 30(1883): 70-152.
RUNGS, C. E. E., 1981.– Catalogue raisonné des Lépidoptères du Maroc. Inventaire faunistique et observations écologiques, .: 365 pp. Traveaux de l’Institut Scientifique, Série Zoologie, n. 40, Rabat.
SCALERCIO, S., INFUSINO, M. & HAUSMANN, A., 2014.– Advances in the knowledge of the Larentiinae fauna of southern Italy by DNA barcoding (Lepidoptera, Geometridae).– Spixiana, 37(2): 260.
SIMOGLOU, K. B., KARATARAKI, A., RODITAKIS, N. E. & RODITAKIS, E., 2012.– Euzophera bigella (Zeller) (Lepidoptera: Pyralidae) and Dasineura oleae (F. Low) (Diptera: Cecidomyiidae): emerging olive crop pests in the Mediterranean?- Journal of Pest Science, 85: 169-177.
SKOU, P. & SIHVONEN, P., 2015.– Ennominae I.– In A. HAUSMANN. The Geometrid Moths of Europe, .: 658 pp. Brill, Leiden.
WIEMERS, M. & FIEDLER, K., 2007.– Does the DNA barcoding gap exist? - a case study in blue butterflies (Lepidoptera: Lycaenidae).– Frontiers in Zoology, 4, 8.